What Is Genetic Testing?
Science • February 23, 2022 • 17min read

Introduction
What if you could take a peek into the future and check on yourself? What if you could know the possible diseases that you may contract in the future? And, what if you could prevent these health problems from occurring in the first place?
Your DNA may hold the secret to that.
Genetic testing or DNA testing is a method to examine the DNA to reveal genetic changes or mutations that predispose a person to certain health risks or disorders. It looks at variations in the sequence of nucleotides, the basic units of the DNA, represented by letters “A” for Adenine, “T” for Thymine, “C” for Cytosine, and “G” for Guanine. It has revealed genetic causes that make certain people more susceptible to diseases as well as guide the development of personalized treatments.
There are several kinds of genetic testing methods that uncover varying depths of information to examine the genome, the full collection of genetic information of an organism. In this article, we’ll discuss these options to know which kinds of methods are fit to be used for different purposes. For simplicity, these can be categorized into single-gene testing, panel testing, and large-scale testing.
I) Single Gene Testing
PCR (Polymerase Chain Reaction)
PCR is a revolutionary method to rapidly create million to billion copies of DNA, allowing scientists to study genes even from very small amounts of samples.
The process has three stages: denaturation, annealing, and extension. During denaturation, the two strands of the DNA helix are separated from each other to serve as templates to amplify the gene of interest. At the annealing stage, a short piece of single-strand DNA called primer is designed to bind specifically to a DNA region where the target gene is located.
Lastly, during extension, when the primer has bound to the desired gene, it acts as a starting point to replicate identical strands of DNA from the original template and create new pieces of DNA.
To create a new DNA, PCR needs the four readily-usable nucleotide forms called dNTP (deoxynucleotide triphosphates) as building blocks to create all the new DNA strands:
- deoxyadenosine triphosphate (dATP)
- deoxythymidine triphosphate (dTTP)
- deoxycytidine triphosphate (dCTP)
- deoxyguanosine triphosphate (dGTP)
Additionally, a magnesium-dependent enzyme called DNA polymerase is also required to perform the actual DNA extension using those dNTPs to grow the new DNA. The whole cycle is repeated 20-40 cycles to achieve exponential amplification in just around 2 hours.
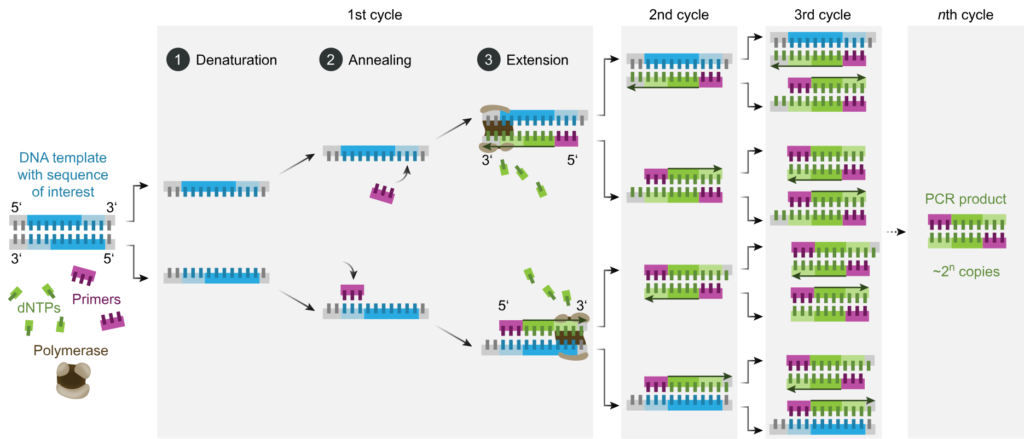
Image source: Wikipedia
After producing large amounts of DNA, visualization through fluorescent staining is used to qualitatively detect if a human gene or a pathogen gene is present or absent. This technique became a major method in molecular biology research and a foundation of more advanced genetic technologies.
RT-PCR (Reverse Transcription-Polymerase Chain Reaction)
RT-PCR is a variation of the PCR technology. Unlike qualitative PCR (results show the presence or absence of genes), RT-PCR is quantitative. It is also designed to target specific genes, but its main purpose is to determine the expression levels of a gene.
In this method, the starting material is mRNA (messenger RNA), the single-stranded form of genetic material, produced in variable quantities during gene expression when the gene is turned on/off. Using a process called reverse transcription, mRNA is first converted to cDNA (complementary DNA) to make it double-stranded so that it can undergo the PCR cycles of denaturation, annealing, and elongation.
The body has different kinds of cells which express different sets of genes to perform specific functions. There are also universal genes that are expressed stably and constantly at all times in any kind of cell in the body that play critical roles in basic cellular processes and survival and these are called housekeeping genes.
Due to its constant, stable, and universal nature in all cells, housekeeping genes are used in RT-PCR experiments as quality controls or standards that serve as expression level references for the measurement of other genes.
Two widely-used housekeeping genes in RT-PCR are GAPDH (Glyceraldehyde 3-phosphate dehydrogenase), a key molecule that facilitates the conversion of food to energy during a process called glycolysis, and β-actin, structural protein needed by cells to retain its shape, integrity, and motility.
In interpreting RT-PCR results, there is an important parameter called the cycle threshold (Ct) value. As an example, for infectious disease testing, like in COVID-19, the resulting Ct value used to test for a SARS-CoV-2 gene can determine if a person is positive for the disease, as well as correlate with viral load, infectivity, disease severity, and mortality. The Ct value is inversely proportional to the gene expression level – that is to say, the lower the Ct value, the higher the viral load, and vice versa.
From the starting viral cDNA, the genes will be amplified exponentially for up to 40 cycles. Then, the continuous multiplication of viral genes will be recorded per cycle by the machine and visualized as a curved graph of fluorescence vs cycle number.
The curve can show the Ct value which is the cycle number at which the fluorescent signal is detected as a threshold of positivity, or basically, it’s the point at the curve at which the amplification starts to be exponential.
Suppose the Ct value is determined during early cycles, in that case, it means that the amount of viral genetic material is too high that it does not need too many amplification cycles to generate a signal to be detected. In the same way, if viral load is too low, it will need many cycles to generate a detectable signal, thus informing if the patient’s clinical sample contains significant viral RNA.
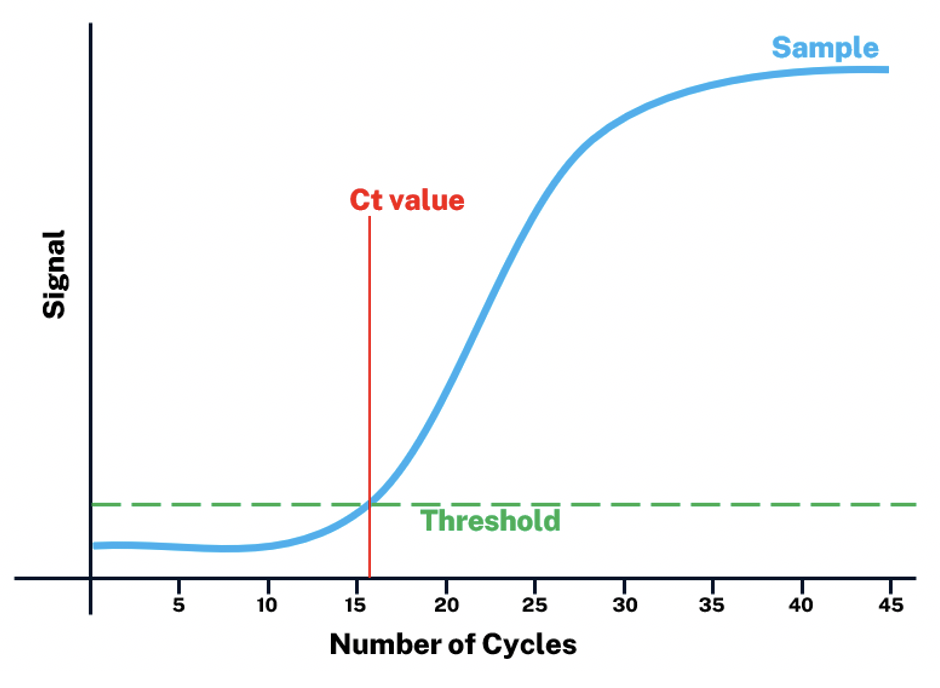
Image source: albany.edu
Sanger Sequencing
Sanger sequencing is the first sequencing technology to sequence the order of nucleotides of genes.
Sanger sequencing uses PCR and chain-termination techniques. In this method, the nucleotides themselves are labeled with specific fluorescent dyes such that A, T, C, and G have their signal. So, during elongation of the new DNA strand, each time a single fluorescent nucleotide is added, the chain terminates, thereby creating numerous fragments of different sizes spanning the length of the gene.
These fragments will be separated according to sizes, and then their fluorescent signals will be visualized as a wave graph called chromatogram where the peaks represent the signals of specific nucleotides and are recorded by the machine to conclude the actual genetic sequence.
Many types of mutations can happen in a gene that can affect its structure and function. Depending on what type of genetic change or mutation happened and where it is in the sequence, the gene may become shorter/longer, non-functional, less/more active, may perform new functions, or have no effect at all.
Through sequencing, mutations in genes can be spotted and computational analyses can provide insights into possible consequences on health and disease.
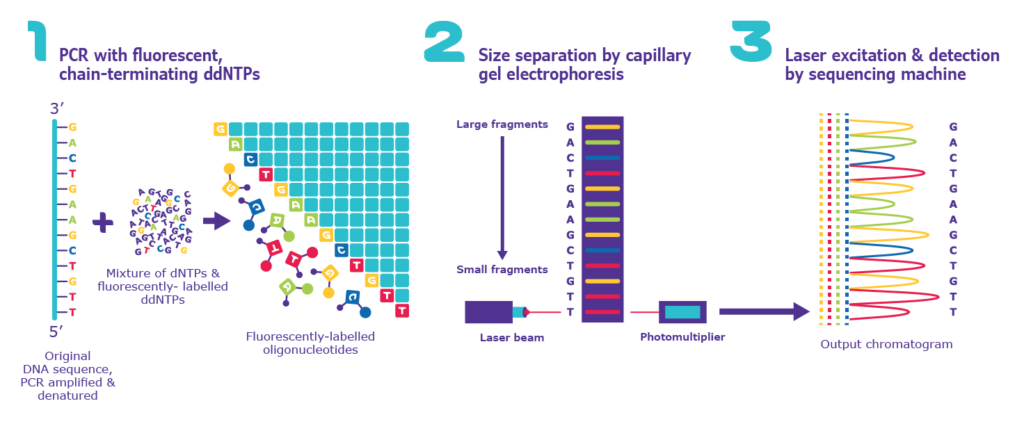
Image source: sigmaaldrich.com
II) Panel Testing
DNA Microarray Testing
Microarray Testing does not sequence the full gene length, but specifically identifies the genetic changes present in predefined regions of the DNA only, allowing testing for genetic variants called SNPs (single-nucleotide polymorphisms). As the name suggests, SNPs are the different forms of nucleotides present in a specific single site in a gene.
For example, in a hypothetical gene sequence TACCCTAATGCGACG, the majority of people may have the original gene sequence TACCCTAATGCGACG (variant 1), while a subset of the population may have TACCCTAACGCGACG (variant 2) – the same gene, two variants, but only differs in a single nucleotide in the sequence.
Here, we can say that there is an SNP at position 9 of the gene, harboring a “T” or “C”. Depending on the nucleotide type and its location in the gene sequence, it may affect the gene expression, structure, and function of the molecule it encodes, impacting traits and diseases.
In microarray testing, it uses an SNP array in a biochip where a large number of tiny spots are attached to its surface. Each spot contains variant-specific DNA probes (sequence-specific oligonucleotides or short stretches of DNA) designed to bind to target DNA regions or genes.
The sample DNA is first extracted, PCR-amplified, and then labeled with different fluorescent dyes. When the target genes are bound to the SNP arrays, a specialized scanning software scans the fluorescence to detect the signals and record the detected SNP.
SNP Microarray is the technology behind direct-to-consumer (DTC) or at-home genetic testing services that are now available in the market. Using just saliva or a buccal swab, anyone can avail of genetic testing services from companies to look at SNPs correlated with lots of risks and traits, including ancestry.
Some of the tested SNPs are correlated with nutrition, fitness, personality, cognition, diseases, skincare, and longevity. The companies also recommend lifestyle or prophylactic changes as well as design DNA-based supplements to optimize health and vitality.
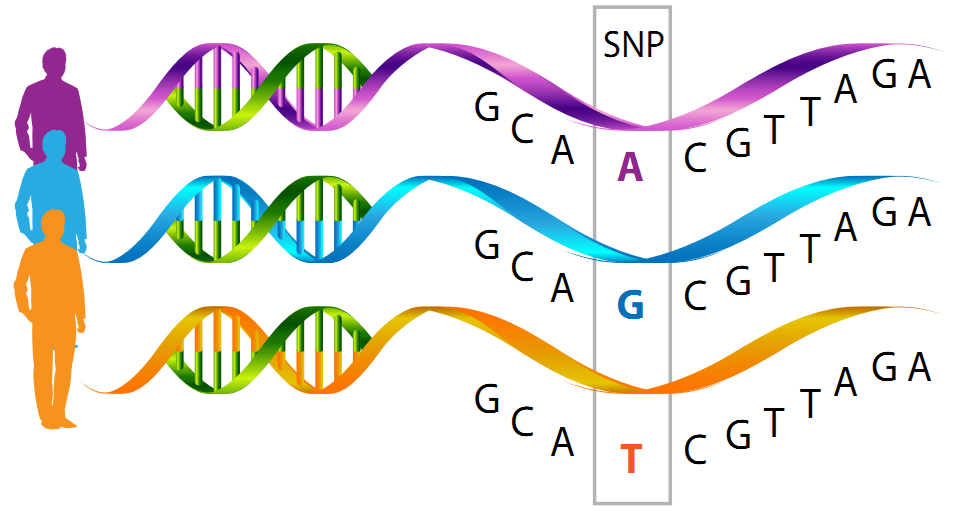
Image source: nutrigeneticsspecialists.com
Targeted Sequencing
Human DNA has about 20,000 genes and 3 billion nucleotides. Sequencing everything is too expensive and generates enormous data that may not be needed for disease-specific investigations. Targeted sequencing is a cost-effective method focusing on specific areas in the genome to sequence only a desired number of genes.
Therefore, targeted genes may be customized for addressing unique research interests or maybe pre-designed by companies to particularly provide the known or established disease- or trait-related genes only.
For sequencing small numbers of genes, Sanger Sequencing can be sufficient, but for a large number of desired targets, a Next-Generation Sequencing (NGS) panel containing hundreds to thousands of disease/trait-specific primers is needed to identify the sequences.
NGS technologies are those that employ high-throughput DNA sequencing approaches by applying massively parallel processing (the utilization of a large number of computer processors to simultaneously perform computations in parallel with each other). Aside from Targeted Sequencing, examples of NGS include Whole-Genome Sequencing and Whole-Exome Sequencing.
Typically, NGS methods have four major steps:
- Library preparation – DNA is fragmented and then oligonucleotides are attached to the two ends of DNA fragments, simultaneously, followed by PCR and purification. The resulting collection of modified DNA fragments is called a DNA library
- Cluster Generation – DNA library is dispensed to the flow cell (a small reactor plate where all the sequencing happens) which contains oligos (short stretches of DNA) on its surfaces to capture the complementary oligonucleotides of the DNA library, then it will undergo bridge amplification cycles in the flow cell to create clusters of same fragments
- Sequencing – In the clusters, fluorescently-labeled nucleotides are elongated complementary to the template DNA fragments. As each nucleotide is added to the growing strand, the fluorescent signals in each cluster are read and recorded
- Data Analysis – Lastly, computer programs allow the full DNA sequence to be reconstructed by aligning it to a reference genome
In Targeted Sequencing, two popular methods used are Hybridization Capture and Amplicon Sequencing, which mainly differ in the method by which samples are amplified before the actual sequencing step. For Hybridization Capture, biotin-labeled oligonucleotides are used as DNA probes to capture the target genes in the DNA library.
On the other hand, Amplicon Sequencing utilizes DNA primers and PCR for simultaneous amplification of multiple genes. Then, the sequencing and data analysis steps are the same.
![]()
Image source: genscript.com
III) Large-Scale Testing
Whole Exome Sequencing (WES)
DNA is a nucleotide string of genes composed of protein-coding regions (exons) and non-coding regions (introns). But, for a gene to be functional, the exons are joined together to be converted to mRNA and then later on to a protein, while the intron parts are removed.
Most known mutations occur in exons because they code for proteins that actively function and support the activities inside the cells. Therefore, the mutations in exons may cause direct impacts on the development of abnormalities and disorders. True enough, exons contain around 85% of all the previously-known variants correlated to diseases.
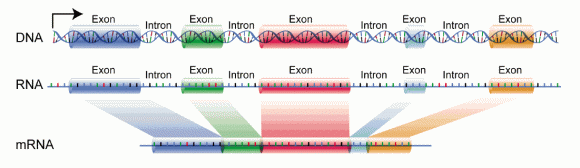
Image source: differencebetween.com
As another NGS method, this WES is designed to sequence the exome – the collection of all exons. Sequencing only the exome provides broad coverage of genes and may be more cost-effective and data-efficient than doing full-blown whole-genome sequencing.
WES is widely-used for cancer applications. Since cancer is caused by mutations that make genes abnormally high or deficient, WES has been instrumental to identify which genes are those that became aberrant and support tumor progression.
This information can be utilized to diagnose what kind of cancer a patient has, what stage it is, identify mortality risk, and even find the exact drugs that therapeutically target those genes. In cancer immunotherapeutics, these forms of mutated proteins are called neoantigens.
These neoantigens can be recognized by the immune system as a search-and-destroy mechanism of natural immune defense and can be further developed into a cancer vaccine to help the body fight tumors. Neoantigen identification can be done typically by using WES.
Whole Genome Sequencing (WGS)
When the first human genome was sequenced, the cost was around US$3 billion and it took 20 years to complete in 2001 using Sanger Sequencing technology. In the modern-day, WGS can do this for just US$ 3,000-5,000 and a recent breakthrough in rapid genome sequencing can do it in just 5 hours. WGS is also an NGS method that comprehensively and completely sequences the full order of all nucleotides in the genome.
Initially, research was focused on exons only, and introns were treated as junk DNA. However, through time, scientists realized the importance of introns in regulating gene functions. Therefore, investigating the full genomes through WGS became critical to finding genetic variants in any part of the genome and understanding the genetics of life from a broader perspective.
In clinics and hospitals, WGS is used for diagnostics of genetic disorders by completely scanning for mutations anywhere in the genome. For example, doctors can identify what mutations can cause a gene to malfunction resulting in a disorder.
WGS is particularly useful in diagnosing rare diseases. Many children with rare diseases took several years and some even a decade for their conditions to be diagnosed, which was only until WGS was done. Since some rare diseases can be caused by novel mutations that have not been previously characterized, WGS can easily find the causal variants because of its full sequencing coverage of the whole genome as opposed to other NGS methods.
In general, novel (newly-discovered) mutations are identified through next-generation sequencing. When a patient’s DNA is sequenced, the sequences will be aligned to a control/standard/reference genome representing a healthy individual.
All the usual nucleotides in the reference genome have already been characterized throughout the years, therefore, the variants or alleles – the possible nucleotide variants that should be present in each position in the DNA sequence – for a healthy individual are already known.
Hence, during computational data analysis, if the results show that a patient harbors a genetic variation that is not usually present in the normal human population or exists only rarely, it is classified as a mutation. This mutation will further be studied if it can affect the structure or function of the gene and if it impacts the development or progression of a disease.
When the genetic mutation is diagnosed, doctors and scientists can design a specific drug or tailor therapeutic strategies to ameliorate the disorder or prevent toxicities.
Risks And Limitations
As with any technology, there are also risks and limitations including accuracy and regulation. accessibility, ethnicity, variants of unknown significance, data privacy, and emotional concerns.
In terms of accuracy and regulation, diagnostic NGS methods offered in accredited clinics and hospitals provide higher confidence in genetic testing. These diagnostic genetic tests that are used for moderate to high-risk purposes (e.g. diagnosing a disease) are required to undergo FDA review and approval to evaluate if it is accurate and valid to serve their claimed purpose.
On the other hand, the non-diagnostic and DTC testing that are used for non-medical, general wellness, or low-risk medical purposes (e.g. ancestry, health risks/predispositions, trait likelihoods in diet and exercise) do not need FDA review before it can be offered to the public.
As such, the accuracy in the former is higher than in the latter. Since the FDA doesn’t assess the clinical accuracy and validity of the claims in the latter, the insights may be limited; however, it may be enough for estimating general risks as these tests are also based on peer-reviewed studies.
Peer-reviewed studies are research works that are accomplished by experts and are also reviewed by other experts in the same field before they can be published in journals to make sure that the information, methods, and findings are scientifically correct or valid following the journals’ standards and process.
At the start, scientists first conduct a research investigation and write the full study details and results as an article, then it will be sent to the journal editor for initial checking. The editor will then send it to anonymous expert reviewers for evaluation, feedback, and suggestions. If the article meets the editorial and peer standards, it will be published in the journal, but if not, it may be returned to the authors for revision or it may also be rejected.
This way, by using the discoveries that are only from peer-reviewed studies for actual health applications, the design, and development of genetic testing services are backed by reliable, high-quality, and strong scientific evidence to ensure the accuracy and validity of non-diagnostic DTC testing even without FDA review.
DTC testing is also more accessible and cheaper than NGS services. However, DTC tests in different companies are not created equal because there are no standards yet, so if some important variants are not included in the DTC tests, risk estimation may also encounter issues.
Ethnicity also plays a role. Most genetic sequencing or variant studies are also based on European and American populations, so the utility of some variants may not apply to underrepresented populations. Some variants are strongly correlated only for specific ethnic origins.
Certain ethnic groups may have higher risks of developing a disease while other ethnicities do not. For example, the genetic risk for cystic fibrosis is higher for the European population while the genetic risk for Spinal Muscular Atrophy varies across all ancestries.
Variants may be pathogenic (disease-causing) or benign (no effect). There are also variants discovered through sequencing studies that are found to be correlated but still have unknown significance or impact on disease development. Therefore, its interpretation in genetic testing has been challenging.
There is also a concern about genetic data privacy. Since DNA is unique to people and the testing company will have access to it, their privacy may be violated discreetly for illegitimate purposes. Therefore, it’s important to read the terms and conditions of a company before agreeing on their tests.
Depending on the emotional quotient of a person, genetic testing may be able to help them manage disorders with a positive outlook or negatively affect their morale if they get some depressing results from genetic testing. Since genetic risks may also be inheritable, knowing the genetic risks for a person may also provide clues of similar risks to the families.
Conclusion
Overall, the promises of genetic testing provide hope for more advanced diagnostic and therapeutic strategies for many health conditions paving the era of precision medicine – the development of personalized treatment programs based on your DNA.
There are many applications in this field, including newborn screening, forensic testing, paternity testing, ancestry, disease risks, clinical diagnosis, lifestyle, nutrition and weight management, pharmacogenomics, adverse effect prediction, and therapeutics development.
Surely, genetic testing will continue to revolutionize saving lives, and changing how the world cures diseases. Through genetic testing, you may be able to take a peek and gain insights into your future health, prevent diseases from happening, or obtain a drug that is customized only for yourself to get the best treatment in the future.
And your DNA may hold the secret to that.